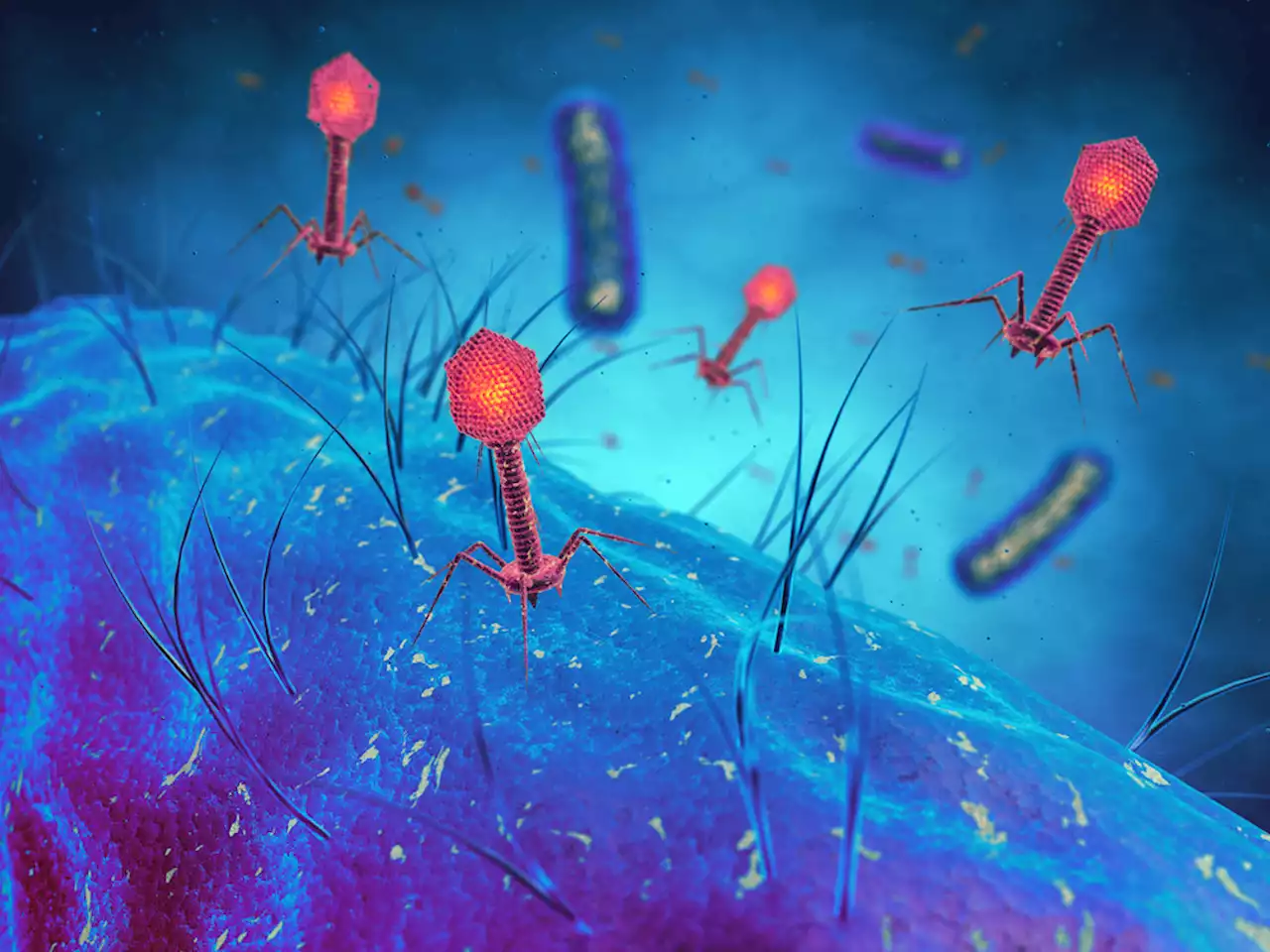
Phages can sense bacterial DNA damage, which triggers them to replicate and jump ship.
, works a bit like HIV. Upon entering the bacterial cell, lambda decides whether to replicate and kill the cell outright, like most viruses do, or to integrate itself into the cell’s chromosome, as HIV does. If the latter, lambda harmlessly replicates with its host each time the bacteria divides.
But, like HIV, lambda is not just sitting idle. It uses a special protein called CI like a stethoscope to listen for signs of DNA damage within the bacterial cell. If the bacterium’s DNA gets compromised, that’s bad news for the lambda phage nested within it. Damaged DNA leads straight to evolution’s landfill because it’s useless for the phage that needs it to reproduce.
México Últimas Noticias, México Titulares
Similar News:También puedes leer noticias similares a ésta que hemos recopilado de otras fuentes de noticias.
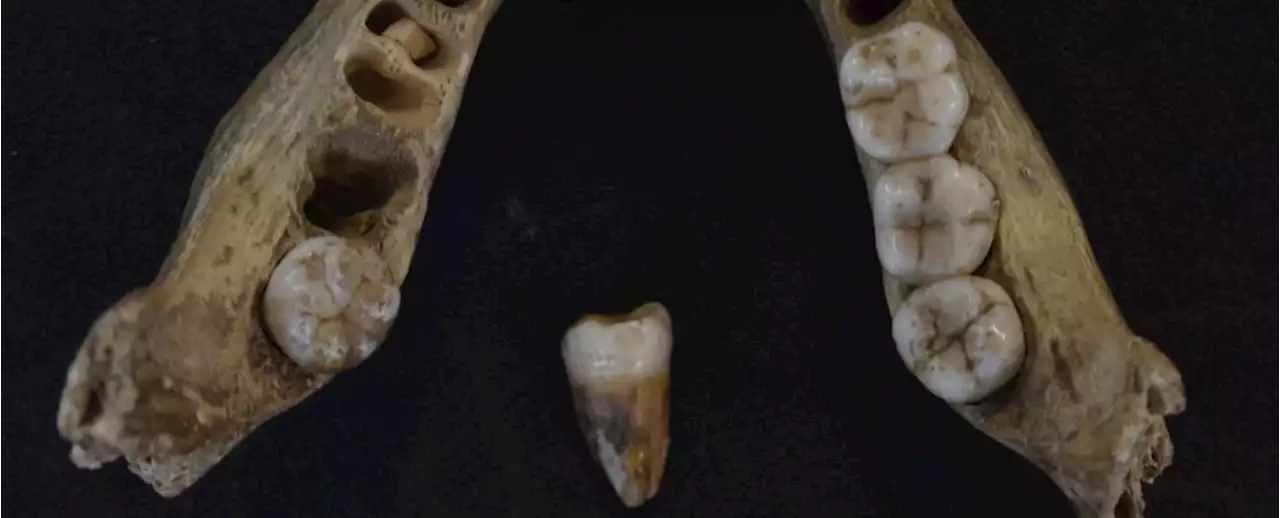 Oldest Human DNA in UK Reveals Ancient Peoples Emerging From The Ice AgeAround 27,000 years ago, an immense sheet of ice coated two-thirds of the British Isles, making the region less than hospitable for human habitation.
Oldest Human DNA in UK Reveals Ancient Peoples Emerging From The Ice AgeAround 27,000 years ago, an immense sheet of ice coated two-thirds of the British Isles, making the region less than hospitable for human habitation.
Leer más »
 Exhumations resume for DNA to ID Tulsa Race Massacre victimsSome of the 19 bodies exhumed for testing in an effort to identify victims of the 1921 Tulsa Race Massacre and then reburied in an Oklahoma cemetery will be removed again starting Wednesday to gather more DNA.
Exhumations resume for DNA to ID Tulsa Race Massacre victimsSome of the 19 bodies exhumed for testing in an effort to identify victims of the 1921 Tulsa Race Massacre and then reburied in an Oklahoma cemetery will be removed again starting Wednesday to gather more DNA.
Leer más »
 Exhumations resume for DNA to ID Tulsa Race Massacre victimsAt a cemetery in Tulsa, Oklahoma, bodies are being removed from their graves for a second time in an effort to identify victims of the Tulsa Race Massacre. tulsa tulsaracemassacre kprc2 click2houston
Exhumations resume for DNA to ID Tulsa Race Massacre victimsAt a cemetery in Tulsa, Oklahoma, bodies are being removed from their graves for a second time in an effort to identify victims of the Tulsa Race Massacre. tulsa tulsaracemassacre kprc2 click2houston
Leer más »
 Exhumations resume for DNA to ID Tulsa Race Massacre victimsHistorians say the white mob violence in Tulsa that targeted Black Americans in 1921 left between 75 and 300 people dead.
Exhumations resume for DNA to ID Tulsa Race Massacre victimsHistorians say the white mob violence in Tulsa that targeted Black Americans in 1921 left between 75 and 300 people dead.
Leer más »
 Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains - Nature BiotechnologyTargeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains
Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains - Nature BiotechnologyTargeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains
Leer más »
![]() Microbial Tracking-2, a metagenomics analysis of bacteria and fungi onboard the International Space Station - MicrobiomeBackground The International Space Station (ISS) is a unique and complex built environment with the ISS surface microbiome originating from crew and cargo or from life support recirculation in an almost entirely closed system. The Microbial Tracking 1 (MT-1) project was the first ISS environmental surface study to report on the metagenome profiles without using whole-genome amplification. The study surveyed the microbial communities from eight surfaces over a 14-month period. The Microbial Tracking 2 (MT-2) project aimed to continue the work of MT-1, sampling an additional four flights from the same locations, over another 14 months. Methods Eight surfaces across the ISS were sampled with sterile wipes and processed upon return to Earth. DNA extracted from the processed samples (and controls) were treated with propidium monoazide (PMA) to detect intact/viable cells or left untreated and to detect the total DNA population (free DNA/compromised cells/intact cells/viable cells). DNA extracted from PMA-treated and untreated samples were analyzed using shotgun metagenomics. Samples were cultured for bacteria and fungi to supplement the above results. Results Staphylococcus sp. and Malassezia sp. were the most represented bacterial and fungal species, respectively, on the ISS. Overall, the ISS surface microbiome was dominated by organisms associated with the human skin. Multi-dimensional scaling and differential abundance analysis showed significant temporal changes in the microbial population but no spatial differences. The ISS antimicrobial resistance gene profiles were however more stable over time, with no differences over the 5-year span of the MT-1 and MT-2 studies. Twenty-nine antimicrobial resistance genes were detected across all samples, with macrolide/lincosamide/streptogramin resistance being the most widespread. Metagenomic assembled genomes were reconstructed from the dataset, resulting in 82 MAGs. Functional assessment of the collective MAGs showed a propen
Microbial Tracking-2, a metagenomics analysis of bacteria and fungi onboard the International Space Station - MicrobiomeBackground The International Space Station (ISS) is a unique and complex built environment with the ISS surface microbiome originating from crew and cargo or from life support recirculation in an almost entirely closed system. The Microbial Tracking 1 (MT-1) project was the first ISS environmental surface study to report on the metagenome profiles without using whole-genome amplification. The study surveyed the microbial communities from eight surfaces over a 14-month period. The Microbial Tracking 2 (MT-2) project aimed to continue the work of MT-1, sampling an additional four flights from the same locations, over another 14 months. Methods Eight surfaces across the ISS were sampled with sterile wipes and processed upon return to Earth. DNA extracted from the processed samples (and controls) were treated with propidium monoazide (PMA) to detect intact/viable cells or left untreated and to detect the total DNA population (free DNA/compromised cells/intact cells/viable cells). DNA extracted from PMA-treated and untreated samples were analyzed using shotgun metagenomics. Samples were cultured for bacteria and fungi to supplement the above results. Results Staphylococcus sp. and Malassezia sp. were the most represented bacterial and fungal species, respectively, on the ISS. Overall, the ISS surface microbiome was dominated by organisms associated with the human skin. Multi-dimensional scaling and differential abundance analysis showed significant temporal changes in the microbial population but no spatial differences. The ISS antimicrobial resistance gene profiles were however more stable over time, with no differences over the 5-year span of the MT-1 and MT-2 studies. Twenty-nine antimicrobial resistance genes were detected across all samples, with macrolide/lincosamide/streptogramin resistance being the most widespread. Metagenomic assembled genomes were reconstructed from the dataset, resulting in 82 MAGs. Functional assessment of the collective MAGs showed a propen
Leer más »